
Centre for Human Genetics
Bangalore.

Bangalore.

Gurudata V Baraka
(Jawaharlal Nehru University, New Delhi),
Centre for Cellular and Molecular Biology,Hyderabad
Post-Doctoral Fellowship, Emory University Atlanta, USA
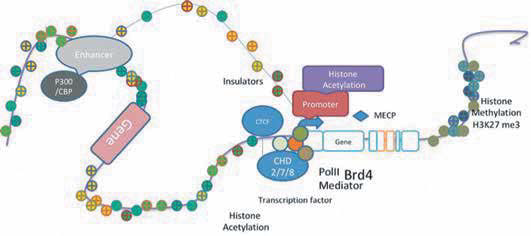
Studies of transcriptional regulation and gene expression in disease phenotypes have guided researchers towards elucidating genetic pathways involved in disease processes. However, such analysis was not always directed at identifying direct targets of transcriptional regulation and gene expression programmes since changes could also be consequences of an earlier event. Also, transcriptional changes could be due to downstream effects of an activated signalling pathway. Techniques were not robust enough to identify relatively modest transcriptional events. Morerecently, the advent of direct RNA sequencing and next-generation sequencing tools have provided us with tools to quantify coding, non-coding andsplicing-variant RNA from the entire genome. For instance, physiological and pathological control of gene expression is often mediated by regulatory proteins or non-coding RNA. DNA-binding sites for regulatory proteins include promoters, enhancers, DNA replication sites and transcription factor targets. A further level of control in the cell is imposed by changes in chromatin structure that could regulate accessibility of these DNA target sequences. To better understand such regulatory events my lab is using disease models along with experimental and bioinformatics approaches for analysis of genome-wide data. It is hoped that this approach will provide a holistic view of the many genes that are likely to be involved in disease processes.he fruit fly Drosophila has proved to be a powerful model system for understanding gene organisation and function, and there is a high degree of optimism over the next phase of disease modelling in fruit flies. As a result, there is growing interest in using flies to explore chromatin contacts and interactions and epigenetics to provide genome-scale descriptions of potential regulatory dysfunctions that result in disease.The hierarchical organisation of the eukaryotic genome is modulated by dynamic interplay of protein-DNA complexes. CCCTC binding factor (CTCF; OMIM 604167) is a DNA-binding protein that is involved in higher-order organisation or chromatin and transcriptional regulation. CTCF is an evolutionarily conserved protein that plays a crucial role in gene regulation as it mediates interchromosomal interactions via chromatin looping. Mutations in CTCF have been reported in patients with intellectual disability and microcephaly. Our study aims to identify the role of CTCF in gene regulation during neuronal development using genetic, molecular and genomic approaches.We are also developing models of autosomal dominant disease conditions by expressing variants of human disease genes (ECZ, BRD4, etc.) in developing Drosophila brain. Epidermolysis bullosa (EB) is a group of inherited skin blistering disorders characterized by spontaneous or trauma-induced blisters on the skin and mucous membranes. It is clinically and genetically heterogeneous. There are four major forms of EB: simplex EB (EBS), junctional EB (JEB), dystrophic EB (DEB) and Kindler syndrome (KS). Definitive diagnosis is based on location of cleavage in the basement membrane zone of the skin. The cleavage occurs within the epidermis in EBS, in the lamina lucida in JEB, and below the lamina densa in DEB. The cleavage plane in KS can be within the basal keratinocytes, lamina lucida or sub-lamina-densa. My lab is working towards creating affordable diagnostic tests for EB by generating all the seven antibodies for EB candidates using in-silico approaches to identify antigenic regions in KRT5, KRT14, laminin alpha, laminin gamma and collagen 7a.
Selected publications:
1.Lamin C and chromatin organization in Drosophila. Journal of Genetics, ;89: 37-49 (2010). Gurudatta BV, Shashidhara LS, Parnaik VK
2.Chromatin insulators: lessons from the fly. Briefings in Functional Genomics, 8: 276-282 (2009). Gurudatta BV, Corces VG
3.The Drosophila transcription factor Adf-1 (nalyot) regulates dendrite growth by controlling FasII and Staufen expression downstream of CaMKII and neural activity. Journal of Neuroscience, 33: 11916-11931 (2013). Timmerman C, Suppiah S, Gurudatta BV, Yang J, Banerjee C, Sandstrom DJ, Corces VG, Sanyal S
4.Dynamic changes in the genomic localization of DNA replication-related element binding factor during the cell cycle.Cell Cycle, 12: 1605-1615 (2013). Gurudatta BV, Yang J, Van Bortle K, Donlin- Asp P, Corces VG
5.The BEAF insulator regulates genes involved in cell polarity and neoplastic growth. Developmental Biology, 369: 124-132 (2012). Gurudatta BV, Ramos E, Corces VG
Copyright © 2011 - All Rights Reserved - Taurus Hard Soft Solutions Pvt. Ltd
Stay Up to Date With Whats Happening